To access the BLAST page in your live window, click on the NCBI icon in the upper left of the page (this takes you to the home page). Click on BLAST in the Popular Resources menu. Carefully read through the list of programs available under "Basic Blast" before proceeding.
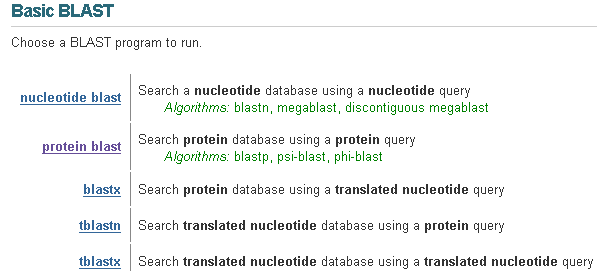
| BLAST Program | Further details |
| nucleotide blast or blastn | Compares a nucleotide query sequence against a nucleotide sequence database. |
| protein blast or blastp | Compares an amino acid query sequence against a protein sequence database. |
| blastx | Compares a nucleotide query sequence translated in all reading frames against a protein sequence database. You could use this option to find potential translation products of an unknown nucleotide sequence. |
| tblastn | Compares a protein query sequence against a nucleotide sequence database dynamically translated in all reading frames. |
| tblastx | Compares the six-frame translations of a nucleotide query sequence against the six-frame translations of a nucleotide sequence database. Please note that the tblastx program cannot be used with the nr database on the BLAST Web page because it is computationally intensive. |
Selecting a BLAST Program:
The "Basic BLAST" menu allows you to do either nucleotide or protein BLAST searches of various types. Because our sequence is a protein sequence, we will do a Protein-protein BLAST (blastp). Click on this option in your live window, then click on NEXT on your tutorial page.
![]()