Part 1: Scroll to the top of your results page and note the Display drop-down box with "GenPept" selected. The GenPept format is the default setting and gives you all of the information we discussed above. However, the FASTA format is more useful for BLAST searches and alignments of sequences. Select FASTA from the menu as illustrated in the screen shot.
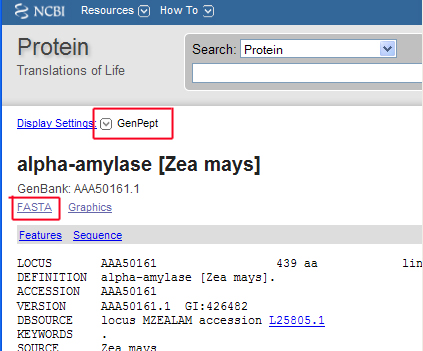
Your results should appear like the screen shot below. You now see less information: just the accession number followed by a brief descriptor, and the amino acid sequence preceded by some identifying information.
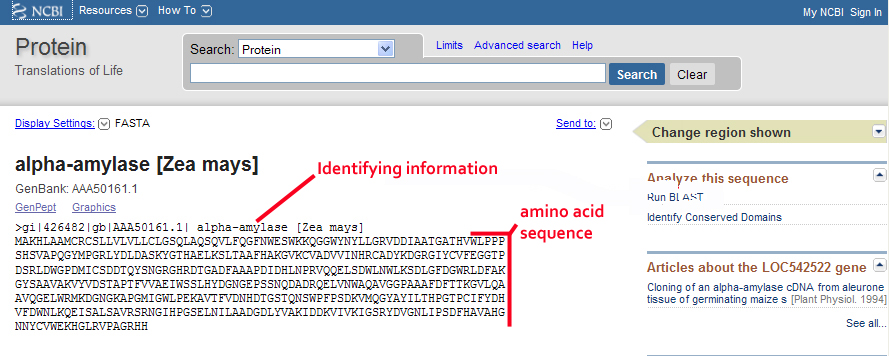
In the live window, highlight and copy the complete amino acid sequence along with the identifying information (>gi|426482…). From your start menu bring up NotePad and paste the FASTA sequence into the window. You will use this sequence in a BLAST search to identify other amino acid sequences in the NCBI databases with similarity to your sequence. Note on this live page that many of the relevant analysis tools that can use this sequence information are linked down the right side of the page. Once you are comfortable using these tools, you can work more efficiently. Minimize NotePad to return to the NCBI website and the tutorial. Click NEXT.